The breeding and genetics group at UGA is active in cutting-edge research projects for the U.S. livestock industry. Our research combines quantitative genetics, genomics, programming, bioinformatics, and statistics. We collaborate with the largest U.S. breeding companies and associations in dairy, beef, swine, poultry, and fish. We have access to extensive data sets across species and premier computing facilities. We also have international contacts with leading research centers worldwide.
The following is a selected list of the current projects. Other projects can benefit from your collaboration. If you are looking for a graduate school or a place for a sabbatical, please consider the University of Georgia.
Joint analysis of phenotype, pedigree and genomic data
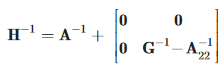
We focused on single-step genomic evaluation. It is BLUP, where the numerator relationship matrix A is replaced by matrix H that combines pedigree and genomic relationships; a lead paper by Aguilar et al. (2010) was chosen as 2013 most cited paper in Genetics and Breeding. The end result of single-step is a dramatic simplification of evaluation procedures combined with superior accuracy, ability to use any model and speed similar to that of BLUP. Single-step is already incorporated in routine evaluations by major genetics companies across species.
A problem in single-step GBLUP (ssGBLUP) was difficulty inverting G with a large number of genotyped animals; more than 1 million Holsteins are genotyped. We developed an algorithm called APY that makes getting the inverse for millions of animals simple, with a greater accuracy than a regular inverse. Read papers by the references on this exciting discovery. Results of APY put a new thinking on limits of genomic selection and on GWAS, with potential for many exciting papers.
Selected references for ssGBLUP
- Aguilar, I., I. Misztal, D. L. Johnson, A. Legarra, S. Tsuruta, and T. J. Lawlor. 2010. A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J. Dairy Sci. 93:743:752. https://doi.org/10.3168/jds.2009-2730
- Legarra, A., I. Aguilar, and I. Misztal. 2009. A relationship matrix including full pedigree and genomic information. J. Dairy Sci. 92:4656-4663. https://doi.org/10.3168/jds.2009-2061
- Lourenco, D. A. L., I. Misztal, S. Tsuruta, I. Aguilar, E. Ezra, M. Ron, A. Shirak, J. I. Weller. 2014. Methods for genomic evaluation of a relatively small genotyped dairy population and effect of genotyped cow information in multiparity analyses. J. Dairy Sci. 97:1742–1752. https://doi.org/10.3168/jds.2013-6916
- Misztal, I., A. Legarra, and I. Aguilar. 2009. Computing procedures for genetic evaluation including phenotypic, full pedigree and genomic information. J. Dairy Sci. 92:4648-4655. https://doi.org/10.3168/jds.2009-2064
- Tsuruta, S., I. Aguilar, I. Misztal, and T. J. Lawlor. 2011. Multiple-trait genomic evaluation of linear type traits using genomic and phenotypic data in US Holsteins. J. Dairy Sci. 94:4198-4204. https://doi.org/10.3168/jds.2011-4256
Selected references for ssGBLUP with APY
- Fragomeni, B. O., D. A. L. Lourenco, S. Tsuruta, Y. Masuda, I. Aguilar, A. Legarra, T. J. Lawlor, and I. Misztal. 2015. Use of genomic recursions in single-step genomic BLUP with a large number of genotypes. J. Dairy Sci. 98:4090-4094. https://doi.org/10.3168/jds.2014-9125
- Lourenco, D. A. L., S. Tsuruta, B. O. Fragomeni, Y. Masuda, I. Aguilar, A. Legarra, J. K. Bertrand, T. S. Amen, L. Wang, D. W. Moser, and I. Misztal. 2015. Genetic evaluation using single-step genomic BLUP in American Angus. J. Anim. Sci. 93:2653-2662. https://doi.org/10.2527/jas.2014-8836
- Masuda, Y., I. Misztal, S. Tsuruta, A. Legarra, I. Aguilar, D. Lourenco, B. Fragomeni and T. L. Lawlor. 2016. Implementation of genomic recursions in single-step genomic BLUP for US Holsteins with a large number of genotyped animals. J. Dairy Sci. 99:1968-1974. https://doi.org/10.3168/jds.2015-10540
- Misztal, I. 2016. Inexpensive computation of the inverse of the genomic relationship matrix in populations with small effective population size. Genetics 202:411-409. https://doi.org/10.1534/genetics.115.182089
- Misztal, I., A. Legarra, and I. Aguilar. 2014. Using recursion to compute the inverse of the genomic relationship matrix. J. Dairy Sci. 97:3943-3952. https://doi.org/10.3168/jds.2013-7752
- . 2016. The dimensionality of genomic information and its effect on genomic prediction. Genetics 203:573-581. https://doi.org/10.1534/genetics.116.187013
See more references in the publications section.
Heat stress in dairy/beef cattle and pigs
Wherever heat stress affects livestock, for example in Southeastern USA, some animals perform satisfactorily but some perform poorly. We developed a methodology to study genetics of heat tolerance using easily available weather records. Our studies indicate that continued selection for performance in moderate climates makes cattle less heat tolerant for production and particularly reproduction. Analyzes of US data for Holsteins indicated negative trends for heat tolerance. Models from dairy are adopted in pigs and beef cattle for growth and for fertility.
Selected references
- Aguilar, I., I. Misztal, S. Tsuruta. 2009. Genetic components of heat stress for dairy cattle with multiple lactations. J. Dairy Sci. 92:5702–5711. https://doi.org/10.3168/jds.2008-1928
- Fragomeni, B. O., D. A. L. Lourenco, S. Tsuruta, S. Andonov, K. Gray, Y. Huang, and I. Misztal. 2016. Modeling response to heat stress in pigs from nucleus and commercial farms in different locations. J. Anim. Sci. 94:4789-4798. https://doi.org/10.2527/jas.2016-0536
- Bradford, H. L., B. O. Fragomeni, D. A. L. Lourenco, and I. Misztal. 2016. Genetic evaluations for growth heat tolerance in Angus beef cattle. J. Anim. Sci. 94: 4143–4150. https://doi.org/10.2527/jas.2016-0707
- Oseni, S., I. Misztal, S. Tsuruta, R. Rekaya. 2004. Genetic components of days open under heat stress. J. Dairy Sci. 87:4327–4333. https://doi.org/10.3168/jds.S0022-0302(04)73434-7
- Ravagnolo, O., I. Misztal, G. Hoogenboom. 2000. Genetic component of heat stress in dairy cattle, development of heat index function. J. Dairy Sci. 83:2120–2125. https://doi.org/10.3168/jds.S0022-0302(00)75094-6
Efficient yet simple animal-breeding programming in modern Fortran

Use of object-oriented and matrix operations in Fortran 90/95/2003/2008 can lead to programs that are almost as simple as in a matrix language but much more efficient. Read a paper titled: Complex models, larger data, simpler computing? This project has resulted in a large number of application programs. Why not C/C++/Java? We focus on algorithms that can be programmed in any language. For numerical computations, modern Fortran seems to be the most efficient yet reasonably simple.
The programs now fully support genomic models and are capable of a large amount of data while remaining simple. The software become faster with optimized libraries replacing legacy subroutines. Conventional BLUP, REML and Gibbs sampling subroutines have been revised to be more efficient in computing time. Now the software are widely used throughout the world.
Selected references
- Aguilar, I., I. Misztal , A. Legarra , S.Tsuruta. 2011. Efficient computation of genomic relationship matrix and other matrices used in single-step evaluation. J. Anim. Breed. Genet. 128:422-428. https://doi.org/10.1111/j.1439-0388.2010.00912.x
- Masuda, Y., S. Tsuruta, I. Aguilar, and I. Misztal. 2015. Technical note: Acceleration of sparse operations for average-information REML analyses with supernodal methods and sparse-storage refinements. J. Anim. Sci. 93:4670-4674. https://doi.org/10.2527/jas.2015-9395
- Misztal, I. 1999. Complex models, larger data, simpler computing? Interbull Bulletin. 20:33-42. (journal site)
- Misztal, I., S. Tsuruta, T. Strabel, B. Auvray, T. Druet, D. H. Lee, 2002. BLUPF90 and
related programs (BGF90). In: Proc. 7th World Cong. Genet. Appl. Livest. Prod., Communication 28-07. (PDF) - Tsuruta, S., I. Misztal and I. Strandén. 2001. Use of the preconditioned conjugate gradient algorithm as a generic solver for mixed-model equations in animal breeding applications. J. Anim. Sci. 79:1166-1172. https://doi.org/10.2527/2001.7951166x
